the drinking water microbiome
|
Microbial control in the drinking water sector is largely oriented towards inactivating, removing, and/or starving microorganisms with the key goal of pathogen elimination. Yet, drinking water systems host a complex consortium of bacteria, archaea, eukaryotes, and viruses collectively referred to as the “drinking water microbiome. We are by the vision to transform the biological paradigm in the drinking water sector from that of microbial elimination to one of microbial exploitation where the natural metabolic potential of the drinking water microbiome is exploited for beneficial purposes. We are engaged in a long-term effort to catalyze this paradigm shift by (1) systematically characterizing the drinking water microbiome, (2) elucidating how engineering decisions and environmental conditions shape it and why, and (3) developing modeling approaches to predict its spatial-temporal dynamics within the current drinking water management paradigm. Cumulative advances in all three themes is critical to effectively attract and engage the water industry, regulators, and consumers in the future of microbial management in the drinking water sector, i.e., proactively engineering a beneficial drinking water microbiome.
|
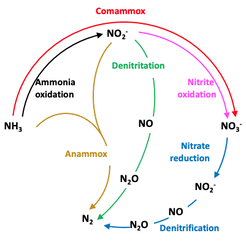
nitrifiers in the engineered water cycle
|
Aerobic nitrification is a globally important nitrogen biotransformation pathway. For more than a 100-years, the aerobic oxidation of ammonia to nitrate was considered to be driven by a collaboration between ammonia and nitrite oxidizers. The discovery of complete ammonia-oxidizing (comammox) bacteria capable of completely oxidizing ammonia to nitrate upended this 100-year-old dogma. Our group was among the first to publish on complete ammonia-oxidizing bacteria. Since then, we are working towards (1) elucidating the ecology of comammox bacteria in engineered systems and (2) their potential applications to improve the sustainability of nitrogen removal from wastewater1. Below are two recent outputs from each theme that substantially advance the state of knowledge of comammox bacteria and their potential applications.
|
low cost real-time microbiome characterization
|
Microbiome data acquisition is a multi-stage process with advanced expertise and infrastructure requirements which limits routine use (e.g., process monitoring and control, diagnostics) and also slows the process of scientific discovery. Can we get more (even if noisy) data rapidly and cheaply to characterize microbial communities? Can we leverage developments in frugal science to minimize infrastructure and expertise requirements? We are working to make nucleic acid sequencing and optical sensing based approaches broadly accessible.
|